PepSite
A structural method to predict peptide/protein bindingEvangelia Petsalaki, Alexander Stark, Eduardo Garcia Urdiales, Rob Russell
Examples
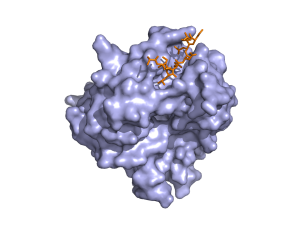
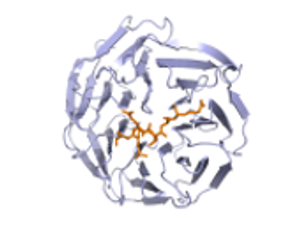
Abstract from Kanelis, V. et al, Nat.Struct.Biol. v8 pp. 407-12, 2001
Nedd4 is a ubiquitin protein ligase composed of a C2 domain, three (or four) WW domains and a ubiquitin ligase Hect domain. Nedd4 was demonstrated to bind the epithelial sodium channel (alphabetagammaENaC), by association of its WW domains with PY motifs (XPPXY) present in each ENaC subunit, and to regulate the cell surface stability of the channel. The PY motif of betaENaC is deleted or mutated in Liddle syndrome, a hereditary form of hypertension caused by elevated ENaC activity. Here we report the solution structure of the third WW domain of Nedd4 complexed to the PY motif-containing region of betaENaC (TLPIPGTPPPNYDSL, referred to as betaP2). A polyproline type II helical conformation is adopted by the PPPN sequence. Unexpectedly, the C-terminal sequence YDSL forms a helical turn and both the tyrosine and the C-terminal leucine contact the WW domain. This is unlike other proline-rich peptides complexed to WW domains, which bind in an extended conformation and lack molecular interactions with residues C-terminal to the tyrosine or the structurally equivalent residue in non-PY motif WW domain targets. The Nedd4 WW domain-ENaC betaP2 peptide structure expands our understanding of the mechanisms involved in WW domain-ligand recognition and the molecular basis of Liddle syndrome.
If you give to pepsite as input the peptide sequence GTPPPNYDSL, the pdbid 1i5h and the chain id W you will get the result linked here.Don't forget to provide a title for the job. Providing an Email might make it easier to track the job later.
2joc : RNEDD4 WWIII Domain from Rattus norvegicus
The PDB file with id 2joc is the equivalent unbound structure
If you give to pepsite as input the peptide sequence GTPPPNYDSL, the pdbid 2joc you will get the result linked here.Don't forget to provide a title for the job. Providing an Email might make it easier to track the job later.
The binding site that PepSite predicts differs from the one shown in the complex, which resides in a central cavity of the protein, but is instead inside an exposed aromatic surface lying on one of the fibronectin domains of MMP2. This surface resembles that for many other proline-rich peptide binding proteins (e.g. SH3, EH1, etc.), and was originally suggested to be the binding site for gelatin based on an early single domain structure (Pickford et al, Structure. 1997 Mar 15;5(3):359-70.) and alanine scanning mutagenesis in MMP9 (Banyai et al, Biochem J. 1994 Mar 1;298 ( Pt 2):403-7.) which showed that residues near to our predicted site were important for gelatin binding. Subsequent studies have confirmed this gelatin binding site in MMP2 (e.g. Briknarová et al, J Biol Chem. 2001 Jul 20;276(29):27613-21. Epub 2001 Apr 24.).
If you give to pepsite as input the peptide sequence GPAGPPGA, the pdbid 1eak and the chain id D you will get the result linked here. Don't forget to provide a title for the job. Providing an Email might make it easier to track the job later.
1e88 : FRAGMENT OF THE GELATIN-BINDING DOMAIN OF HUMAN FIBRONECTIN
The PDB file with id 1e88 contains the gelatin binding fragment of fibrinectin without a peptide bound
If you give to pepsite as input the peptide sequence GPAGPPGA, and the pdbid 1e88 you will get the result linked here. Don't forget to provide a title for the job. Providing an Email might make it easier to track the job later.
Abstract from Till et al, Nat Struct Mol Biol. 2007 Oct;14(10):897-903. 2007 Sep 23.
Argonaute (Ago) proteins mediate silencing of nucleic acid targets by small RNAs. In fission yeast, Ago1, Tas3 and Chp1 assemble into a RITS complex, which silences transcription near centromeres. Here we describe a repetitive motif within Tas3, termed the 'Argonaute hook', that is conserved from yeast to humans and binds Ago proteins through their PIWI domains in vitro and in vivo. Site-directed mutation of key residues in the motif disrupts Ago binding and heterochromatic silencing in vivo. Unexpectedly, a PIWI domain pocket that binds the 5' end of the short interfering RNA guide strand is required for direct binding of the Ago hook. Moreover, wild-type but not mutant Ago hook peptides derepress microRNA-mediated translational silencing of a target messenger RNA. Proteins containing the conserved Ago hook may thus be important regulatory components of effector complexes in RNA interference.
If you give to pepsite as input the peptide sequence PDNGTSAWGE, the pdbid 1w9h and the chain id A you will get the result linked here. Don't forget to provide a title for the job. Providing an email might make it easier to track the job later.
If you compare the predictions with the PDB structure 1ytu you can see it is at the binding site of the RNA, as is mention in Till et al. The peptide motif shown to bind was D/N-E/N/-π-S/T-π-A/G-W-G-E and actually two copies of this. As in the webserver only peptides of length upt o 10 residues are permitted we used only the one copy, i.e.PDNGTSAWGE.
1. 1i5h : RNEDD4 WWIII Domain from Rattus norvegicus
WW domains bind [AP]-P-P-[AP]-Y, and/or phosphoserine- phosphothreonine-containing motifs. The PDB file with id 1i5h is a RNEDD4 WWIII Domain bound to a ENaC peptide.Abstract from Kanelis, V. et al, Nat.Struct.Biol. v8 pp. 407-12, 2001
Nedd4 is a ubiquitin protein ligase composed of a C2 domain, three (or four) WW domains and a ubiquitin ligase Hect domain. Nedd4 was demonstrated to bind the epithelial sodium channel (alphabetagammaENaC), by association of its WW domains with PY motifs (XPPXY) present in each ENaC subunit, and to regulate the cell surface stability of the channel. The PY motif of betaENaC is deleted or mutated in Liddle syndrome, a hereditary form of hypertension caused by elevated ENaC activity. Here we report the solution structure of the third WW domain of Nedd4 complexed to the PY motif-containing region of betaENaC (TLPIPGTPPPNYDSL, referred to as betaP2). A polyproline type II helical conformation is adopted by the PPPN sequence. Unexpectedly, the C-terminal sequence YDSL forms a helical turn and both the tyrosine and the C-terminal leucine contact the WW domain. This is unlike other proline-rich peptides complexed to WW domains, which bind in an extended conformation and lack molecular interactions with residues C-terminal to the tyrosine or the structurally equivalent residue in non-PY motif WW domain targets. The Nedd4 WW domain-ENaC betaP2 peptide structure expands our understanding of the mechanisms involved in WW domain-ligand recognition and the molecular basis of Liddle syndrome.
If you give to pepsite as input the peptide sequence GTPPPNYDSL, the pdbid 1i5h and the chain id W you will get the result linked here.Don't forget to provide a title for the job. Providing an Email might make it easier to track the job later.
2joc : RNEDD4 WWIII Domain from Rattus norvegicus
The PDB file with id 2joc is the equivalent unbound structure
If you give to pepsite as input the peptide sequence GTPPPNYDSL, the pdbid 2joc you will get the result linked here.Don't forget to provide a title for the job. Providing an Email might make it easier to track the job later.
2. 1eak : CATALYTIC DOMAIN OF PROMMP-2 E404Q MUTANT from Homo sapiens
The PDB file with id 1eak contains a complex of the catalytic domain of the type IV collagenase (Gelatinase A MMP-2) complexed with an inhibitor peptideThe binding site that PepSite predicts differs from the one shown in the complex, which resides in a central cavity of the protein, but is instead inside an exposed aromatic surface lying on one of the fibronectin domains of MMP2. This surface resembles that for many other proline-rich peptide binding proteins (e.g. SH3, EH1, etc.), and was originally suggested to be the binding site for gelatin based on an early single domain structure (Pickford et al, Structure. 1997 Mar 15;5(3):359-70.) and alanine scanning mutagenesis in MMP9 (Banyai et al, Biochem J. 1994 Mar 1;298 ( Pt 2):403-7.) which showed that residues near to our predicted site were important for gelatin binding. Subsequent studies have confirmed this gelatin binding site in MMP2 (e.g. Briknarová et al, J Biol Chem. 2001 Jul 20;276(29):27613-21. Epub 2001 Apr 24.).
If you give to pepsite as input the peptide sequence GPAGPPGA, the pdbid 1eak and the chain id D you will get the result linked here. Don't forget to provide a title for the job. Providing an Email might make it easier to track the job later.
1e88 : FRAGMENT OF THE GELATIN-BINDING DOMAIN OF HUMAN FIBRONECTIN
The PDB file with id 1e88 contains the gelatin binding fragment of fibrinectin without a peptide bound
If you give to pepsite as input the peptide sequence GPAGPPGA, and the pdbid 1e88 you will get the result linked here. Don't forget to provide a title for the job. Providing an Email might make it easier to track the job later.
3. 1w9h : PIWI PROTEIN FROM ARCHAEOGLOBUS FULGIDUS.
The PDB file with id 1w9h contains a PIWI domain.Abstract from Till et al, Nat Struct Mol Biol. 2007 Oct;14(10):897-903. 2007 Sep 23.
Argonaute (Ago) proteins mediate silencing of nucleic acid targets by small RNAs. In fission yeast, Ago1, Tas3 and Chp1 assemble into a RITS complex, which silences transcription near centromeres. Here we describe a repetitive motif within Tas3, termed the 'Argonaute hook', that is conserved from yeast to humans and binds Ago proteins through their PIWI domains in vitro and in vivo. Site-directed mutation of key residues in the motif disrupts Ago binding and heterochromatic silencing in vivo. Unexpectedly, a PIWI domain pocket that binds the 5' end of the short interfering RNA guide strand is required for direct binding of the Ago hook. Moreover, wild-type but not mutant Ago hook peptides derepress microRNA-mediated translational silencing of a target messenger RNA. Proteins containing the conserved Ago hook may thus be important regulatory components of effector complexes in RNA interference.
If you give to pepsite as input the peptide sequence PDNGTSAWGE, the pdbid 1w9h and the chain id A you will get the result linked here. Don't forget to provide a title for the job. Providing an email might make it easier to track the job later.
If you compare the predictions with the PDB structure 1ytu you can see it is at the binding site of the RNA, as is mention in Till et al. The peptide motif shown to bind was D/N-E/N/-π-S/T-π-A/G-W-G-E and actually two copies of this. As in the webserver only peptides of length upt o 10 residues are permitted we used only the one copy, i.e.PDNGTSAWGE.